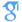Protein–ligand interactions are central to many biological applications, including molecular recognition, protein formulations, and bioseparations. Complex, multisite ligands can have affinities for different locations on a protein’s surface, depending on the chemical and topographical complementarity. We employ an approach based on the spherical harmonic expansion to calculate spatially resolved three-dimensional atomic density profiles of water and ligands in the vicinity of macromolecules. To illustrate the approach, we first study the hydration of model C180 buckyball solutes, with nonspherical patterns of hydrophobicity/-philicity on their surface. We extend the approach to calculate density profiles of increasingly complex ligands and their constituent groups around a protein (ubiquitin) in aqueous solution. Analysis of density profiles provides information about the binding face of the protein and the preferred orientations of ligands on the binding surface. Our results highlight that the spherical harmonic expansion based approach is easy to implement and efficient for calculation and visualization of three-dimensional density profiles around spherically nonsymmetric and topographically and chemically complex solutes.
Reference
Parimal S, Cramer SM and Garde S (). "
Application of a Spherical Harmonics Expansion Approach for Calculating Ligand Density Distributions around Proteins
," J. Phys. Chem. B, 118 (46), 13066-13076
Bibtex
@article{Parimal2014application,
title = {Application of a Spherical Harmonics Expansion Approach for Calculating Ligand Density Distributions around Proteins},
author = {Parimal, Siddharth and Cramer, Steven M and Garde, Shekhar},
journal = {J. Phys. Chem. B},
volume = {118},
number = {46},
year = {2014},
pages = {13066-13076},
doi = {10.1021/jp506849k}
}